Nanostring&冷泉港空間多組學及癌癥研究研討會邀請

胰腺導管癌是一種惡性程度較差的消化系統癌癥,不僅發病隱蔽而且預后極差,預計到2030年它將成為美國第二高致死率的癌種。目前尚無成熟有效的治療方案,醫學界傾向于病人先接受術前治療再進行手術切除來改善預后。然而胰腺導管癌對放化療的抗性以及術前治療對剩余腫瘤細胞可能造成的影響依然是個謎團。在分子分型方面,胰腺導管癌的相關研究也依然處于初級階段。目前通過大塊組織RNA分析手段,只有兩個亞型:classic亞型和basal亞型。而這兩個亞型也尚難對目前的診療方案做出有效的指導作用。因此,通過對胰腺導管癌組織微環境中的癌細胞,免疫細胞和腫瘤相關成纖維細胞進行詳細精準的分子分型和生物標記物分析,將成為改善病人診治和預后的突破口。
近日,麻省理工學院的Aviv Regev團隊首次通過單細胞核RNA測序以及空間全轉錄組學技術的聯合應用,對接受了術前治療和未接受術前治療的胰腺導管癌樣本進行了細胞群落,分子生物標記物和空間信息的深度解析。在胰腺導管癌分子分型深度研究方面,以及由術前治療和免疫微環境對癌細胞造成的影響分析方面獲得了突破性發現。
首先,研究團隊采用單細胞核測序的方法開展細胞分型工作。該方法可避免胰腺導管癌研究的其中一個棘手難題,即由于纖維化和組織降解帶來的細胞損傷對基因表達分析的影響。 然而單純的單細胞核測序并不能把某一特定癌細胞與其周圍富集的免疫細胞或者成纖維細胞進行關聯。因此,研究團隊突破性地采用NanoString的GeoMx DSP空間全轉錄組和空間靶向轉錄組技術,在相同的胰腺導管癌樣本中對組織中不同的細胞群落進行了空間分析,再通過反卷積(Deconvolution)的模型與單細胞核RNA測序的數據結合對映分析,有效分辨出不同癌細胞及其相關的免疫細胞,從而實現在空間信息和單細胞精度層面的最優化。這對新的診療方案開發會更有指導價值。
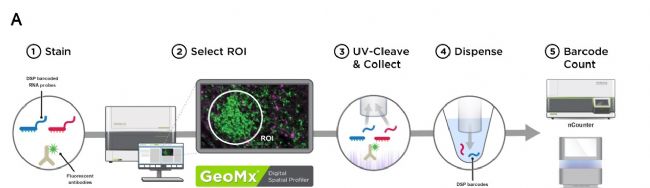
A. Experimental workflow for digital spatial profiling on the GeoMx platform (NanoString)
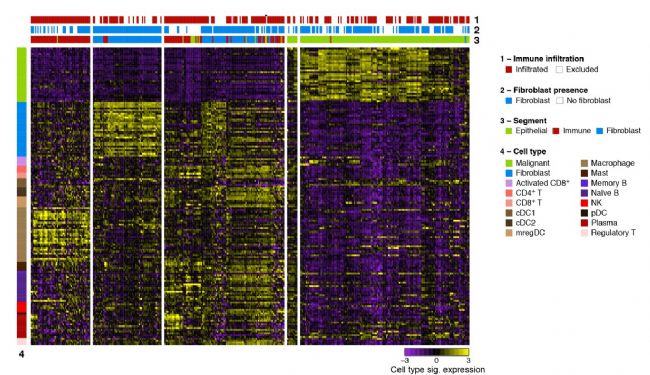
B. Spatial resolution of cell types across ROIs and AOIs. Expression (z-score of normalized counts across AOIs; purple/yellow color bar) of signature genes (rows) from diverse cell types (color legend (4) and left bar) across AOIs (columns, color legend and horizontal bar (3)) profiled by 1,412-gene cancer transcriptome atlas or CTA, capturing epithelial (green), fibroblasts (blue) and immune (red) cells, from ROIs characterized by presence or absence of immune (color legend and horizontal bar (1)) and fibroblast (color legend and horizontal bar (2)) infiltration. Both columns and rows are clustered by unsupervised hierarchical clustering
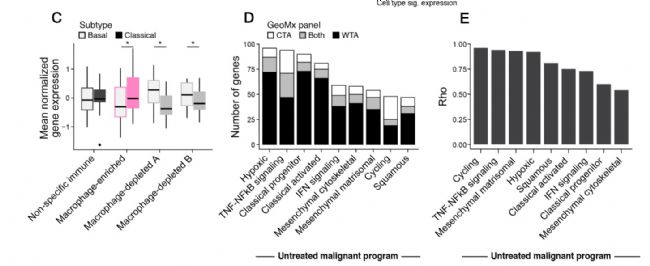
C. Box-plots comparing mean normalized gene expression by cluster for basal-like vs. classical-like epithelial AOIs. * p < 0.05, Student’s t-test.
D.Coverage of PDAC snRNA-seq programs by CTA and WTA digital spatial profiling. Number of genes (y axis) from each untreated malignant cell program (x axis) captured by CTA only (white), WTA only (black), or both (grey)
E. Impact of gene panel on program scores. Spearman correlation coefficient () between the scores for different untreated malignant programs (x axis) obtained with WTA using the full gene panel vs. the gene subset shared with the CTA assay
近年,隨著空間生物學時代的到來,空間轉錄組技術在腫瘤免疫、腫瘤微環境的深度分析應用日新月異。Nanostring GeoMx DSP作為目前腫瘤免疫、微環境研究領域的創新性解決方案和工具,已被國內外多個團隊用于腫瘤,神經科學以及新冠病毒研究。
為使該前沿創新技術不斷助力腫瘤免疫等相關研究,Nanostring將與冷泉港生物科技股有限公司于2020年10月15號下午共同舉辦“空間多組學及癌癥研究技術研討會暨第四屆Nanostring大中華區用戶會”,屆時將邀請中國大陸和臺灣地區的知名腫瘤研究的專家進行技術研討。
會議組誠邀國內專家和客戶踴躍觀看,積極參與會議學術交流。掃描二維碼,成功注冊參會人員,有機會贏取現金紅包。

- 在線講座:BSI全新質譜軟件產品ProteoformX 介紹
- 免費講座:東樂膜片鉗技術系列講座第七期
- 2025領航者峰會邀您一同復盤中國創新藥何以引爆全球
- 講座:利用Cellmatic系統實現hiPSC自動化擴增
- 陶術生物技術講座預告:人工智能藥物設計的實用方法
- 飛納直播:可實現地圖式譜圖加彩色成像的掃描電鏡
- 腦-體互作研究前沿技術培訓會在長沙成功舉辦
- 進科馳安第九場BMG多功能酶標儀線上答疑會邀請函
- CNSD·CNS藥物開發與創新技術發展大會活動議程
- 會議通知:2025年高光譜測量技術及應用學術交流會
- 在線講座:BSI全新版本蛋白質組學高級分析軟件介紹
- MCE直播預告:動物給藥之局部給藥方式及注意事項
- 講座邀請:精準醫學生物信息學前沿研討與高級培訓班
- 瑞沃德系列直播之如何獲取高質量的單細胞懸液
- 免費講座邀請:東樂膜片鉗技術系列講座第六期
- 因美納邀您參加CSCO第九屆血液腫瘤學術大會
- NGDx2025中國先進診斷技術開發與應用論壇議程表
- 10x Chromium GEM-X動物及畜牧類研究獎勵計劃啟動
- Vizgen攜森西賽智亮相空間生物國際會議分享空轉進展
- 怡美通德正式成為PacBio中國區授權代理商
- 相約BioCon2025,因美納邀您共話腫瘤精準醫學新篇章
- 怡美通德邀您共赴單細胞及空間轉錄蛋白多組學論壇
- 10x Genomics技術方案更新(5月),新手冊邀您下載
- 10x新品發布網絡研討會:Visium HD 3’空間分析方案
- Oxford Nanopore明日直播:納米孔測序大會亞太專場
- 10x邀您報名Visium和Xenium數據分析研討會(北京場)
- 第三屆中國國際生殖醫學健康大會(北京站)通知
- 10x Genomics技術方案更新(4月),新手冊邀您領取
- 10x邀您參加單細胞與空間多組學前沿峰會-北京站
- 真邁FASTASeq 300 Dx獲批NMPA三類醫療器械注冊證






